|
||
|
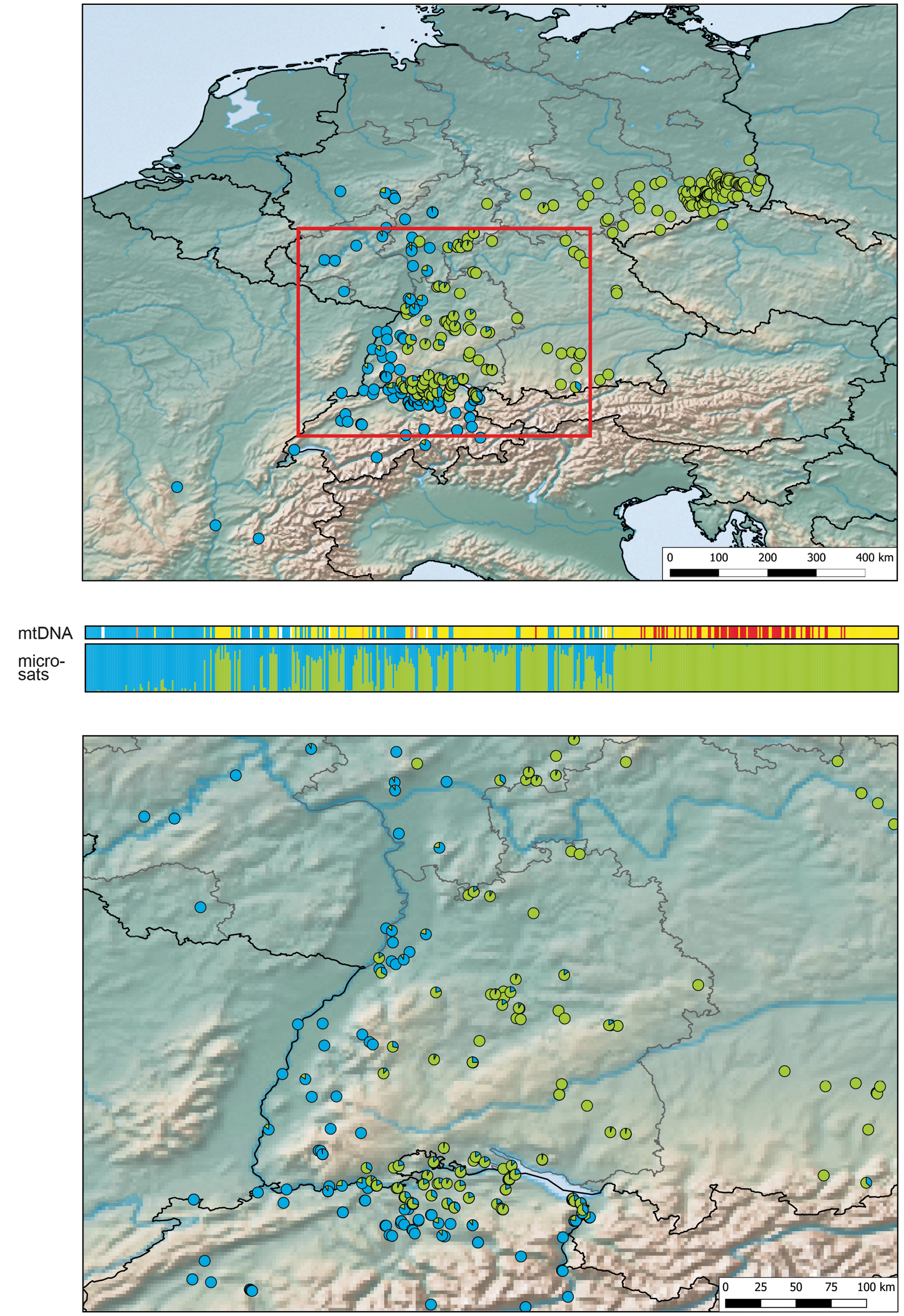
Genotypic identity of grass snakes in the study area. Top: Whole sampling used; blue corresponds to Natrix helvetica; green, N. natrix. The red rectangle highlights the enlarged map segment shown below. Centre: Mitochondrial identity (upper bar) and genotypes (lower bar) of individual grass snakes. Samples are arranged from west to east. The upper bar indicates the mitochondrial haplotype of each individual. Colours represent N. helvetica (blue), the ‘Alpine lineage’ of N. helvetica (orange), the ‘yellow lineage’ of N. natrix (yellow) and the ‘red lineage’ of N. natrix (red). White segments represent missing mtDNA data. STRUCTURE results are shown in the lower bar. There, each vertical column corresponds to an individual snake; colours indicate its genotypic identity for K = 2 (blue = N. helvetica, green = N. natrix). Divided columns with percentages > 5% represent snakes with admixed ancestry. Note that there are many individuals with haplotypes of N. natrix with genomic introgression from N. helvetica, but few snakes harbouring haplotypes of N. helvetica with genomic introgression from N. natrix. Bottom: Detailed map for Baden-Württemberg and adjacent regions. |